Jupyter in the Emacs universe
- 32 minsThe Emacs configurations used to reproduce the screencasts are availabe at github.com/martibosch/snakemacs, under dedicated branches named ein, code-cells-py and code-cells-org respectively (in order of appearance in this blog post). The example notebook with the conda environment required to execute it is available at github.com/martibosch/jupyter-emacs-post.
Jupyter in the Emacs universe
2012: The IPython team released the IPython Notebook, and the world has never been the same
– Jake Vanderplas1
Whether you use Jupyter notebooks or not, it seems hard to disagree with the quote above. I actually did not know that the name “Jupyter” is a reference to the three programming languages to which the IPython notebook was extended in 2014, i.e., Julia, Python and R - again, thank you Wikipedia. By 2018, more than 100 languages were already supported, and quoting Lorena Barba, “For data scientists, Jupyter has emerged as a de facto standard”2. In 2021, Jupyter Notebooks voted by Nature readers as the third software codes that had biggest impact in their work (after the Fortran compiler and the Fast Fourier transform)3.
Unlike Nature and Wikipedia, I would break down Jupyter notebooks into three components (instead of two): the user interface (UI) to edit code and text, the kernel that executes the code and the underlying JSON file format with the “.ipynb” extension into which notebooks are saved and shared. Altogether, the first two components are not much of a novelty since they essentially constitue a read–eval–print loop (REPL) environment, which were developed in the 1960s4. In my view, the novelty is more about how the first two occur within a document-like notebook file editing experience which allows not only to write code cells, but also to move, execute and delete them as desired until the overall look of the notebook is considered satisfactory. At this point, one can save and export the notebook, obtaining a visually-appealing document mixing code, documentation and results, which collectively tells a story, aka, a literate computational narrative5.
However, this editing freedom comes at a non-negligible cost, i.e., the risk of exporting an inconsistent computational pipeline after moving and deleting cells that have been executed (potentially several times). As a matter of fact, in 2019, a study collected a corpus of ~1.16M notebooks from GitHub and found that only 3.17% could be executed providing the same results. Many experienced programmers have raised warnings about how notebooks obscure the state, which can be especially dangerous for beginners6. Additionally, the JSON-based ipynb format of vanilla Jupyter notebooks poses many challenges when it comes to version control, practically requiring you to use external tools such as Jupytext and nbdime to prosper in the face of adversity. But I think that in any case, it is fair to say that, if used properly, Jupyter notebooks are a very powerful tool.
Let us now go back to the first of the three components of notebooks outlined above, namely the interface to edit notebooks. The Project Jupyter provides two options, both in the form of a web application: the classic Jupyter Notebook and JupyterLab, with the latter providing a more fully-featured integrated development environment (IDE) to edit notebooks and other text files, run consoles and manage files. Additionally, many other options have emerged in view of the popularity of Jupyter noteebooks, both web-based (e.g., Google Colab, Azure Machine Learning, GitHub Codespaces, DeepNote…) and client-based (e.g., VSCode, PyCharm, DataSpell…). Nevertheless, for Emacs users such as myself, these are hardly viable options, even when using extensions to emulate Emacs key bindings. Therefore, the goal of this post is to explore which Emacs configurations provide the best Jupyter-like experience.
Desired features
Before we go any further, it is worth noting that this post is inevitably highly opinionanted, or in other words, influenced by the way in which I use Jupyter notebooks, which is twofold. First, like most users, I use notebook to interactively explore, e.g., a dataset, a library or the like. As the notebook code becomes more mature, I may manually move it to Python modules or scripts. Such a development process can lead to both Python libraries or data science repositories. This brings me to the second way in which I use notebooks, namely to tell a story, which can either be the documentation or user guide for a Python library as well as the computational pipeline to reproduce the results of an academic article.
Based on what has been said so far, the main desired features, in no particular order, are the following:
- Beyond (Python) code: mixing Python (or another programming another language), markdown and inline plots can constitute a great recipe to create literate computational narratives, so it is important to ensure that the Emacs setup supports them - otherwise there is practically no reason to use a hardly human-readable JSON-based format instead of plain text “.py” files.
- Proper version control with the possibility to include cell outputs: following the previous point, I am convinced that Jupyter notebooks would not have been this successful if web platforms such as GitHub or GitLab did not offer a rich rendering of notebook files, as reading them directly in the browser can be a very convenient way to navigate computational narratives such as tutorials, computational pipelines to reproduce an academic paper or the like. From this perspective, it is essential that the version-controlled notebooks can include the cell outputs - I emphasize the can, because in some use cases, it may be better to strip out the cell outputs to reduce file sizes and to ease version control, yet it is important that the option to include the outputs exists.
- IDE features (environment-aware and notebook-wide): as highlighted in the introduction, the out-of-order editing and execution of notebooks can dangerously obscure the state, which can be prone to errors and elusive results. Features provided by IDEs such as completion, documentation, on-the-fly syntax checking, navigation, refactoring and the like can help prevent many of these issues. In Emacs, there are several options to provide these IDE features7 - the two main challenges are to ensure that IDE features are aware of the Python environment used in each Emacs buffer, and notebook-wide, i.e., using information (such as imports and variables) defined not only in the cell being edited but in all the notebook cells.
Overview of Emacs configurations for Jupyter notebooks
The following sections assume an overall setup in which multiple virtual Python environments coexist in the same computer. To this end, my configuration8 uses conda (or more precisely, its much faster reimplementation named mamba), with automatic detection and activation of the right conda environment for a buffer using conda.el. Then, Jupyter interacts with conda environments using the ipykernel package9.
Although I believe that conda features major advantages over other virtual environment and package managers10, it should be possible to achieve an analogous setup using alternative tools (e.g., virtualenv and pip11). Similarly, my configuration uses pyright (which actually can be installed using conda)12, a language server protocol (LSP) to obtain IDE features, but feel free to use another tool (e.g., eglot) if you prefer.
Let us now finally move on to experiencing Jupyter notebooks in Emacs.
EIN
The first option is Emacs IPython Notebook (EIN). Using it requires very little Emacs configuration and is quite straightforward: we need to launch a jupyter notebook server (e.g., by running jupyter notebook in a shell or running M-x ein:run in Emacs), then run M-x ein:login. Then, we can either open an existing notebook or select a kernel and create a new notebook. The key bindigns are very convenient and provide a very jupyter-like experience: C-c C-a and C-c C-b respectively create cells above and below the current cell, cells can be moved up and down using M-<up> and M-<down>, C-c C-c executes the current cell, C-c C-w copies the current cell and C-c C-y yanks it, and other bindings allow executing all cells, restarting the kernel and many more. Additionally, you can easily mix Python with markdown, IPython magics, inline plots and shell commands.
Creating a new notebook for a specific Jupyter kernel is also quite straight-forward, i.e., the ein:notebooklist buffer lets you select an option among the existing kernels, and then you just need to click the [New Notebook] button.
While EIN provides an Emacs interface to emulate the web-based Jupyter notebook with its major advantages (i.e., cell organization, markdown rendering, inline results and plots…), such a resemblance comes at the cost of raising several important issues that require careful consideration. First, the “undo” command only works within a given cell, leading to unintuitive undo behaviors when moving accross cells13. Furthermore, a critical aspect to consider is that there is no “undo” for cell deletion, which can easily prompt one to very strongly regret certain typing mistakes. Similarly, Emacs auto-saving and file recovery features do not apply to “ipynb” files edited with EIN, so you can easily lose valuable work if your computer crashes.
Finally, IDE features in EIN are provided using elpy, which provides notebook-wide completion, navigation and documentation, and can be environment aware by using pyvenv-activate or pyvenv-workon. However, based on my experience (and other users have reported similarly), the main issue with elpy is that the background elpy process quickly ends up consiming an entire CPU and provides very slow responses. See a minimal example of how completions and documentation easily become excessively slow:
Moreover, as of May 2023, elpy is unmantained, which I hope that prompts the EIN developers to switch to a LSP client or another package to provide IDE features.
code-cells and emacs-jupyter
Another notable option to make Emacs interact with Jupyter kernels is the emacs-jupyter package. The main difference with EIN is that emacs-jupyter does not provide a graphical UI to edit notebooks. Instead, it just provides the underlying interaction with Jupyter kernels within Emacs, thus giving the user more freedom to chose the notebook editing interface.
The most straight-forward way to use emacs-jupyter is to use the M-x jupyter-run-repl command to run a REPL using a specific Jupyter kernel, open a “.py” file (or whichever language extension corresponds to the Jupyter kernel), write Python code and send the desired lines, regions or the whole buffer to the REPL. While many developers may be perfectly happy with such an approach, it does not represent any major conceptual change with respect to the initial design of the REPL environments in the 1960s. Therefore, it is missing two of the main strengths of Jupyter notebooks, i.e., the organizational compartmentalization provided by notebook cells and the storytelling ability of the exported notebook file, mixing programming code, markdown and inline plots.
In order to emulate the cell-like organization of Jupyter notebooks, code-cells provides a lightweight mode to read, edit and write “.ipynb” files. To that end, it first converts the JSON-based “.ipynb” file to a plain-text script representation, in which certain Python comment syntax is interpreted as cell boundaries. By default, such a conversion is performed by Jupytext (which needs to be installed separately), but it can easily be configured to use any other command (e.g., pandoc) by customizing the code-cells-convert-ipynb-style. In fact, it is also possible to convert “.ipynb” files to another format such as markdown or org, and then edit them in the associated Emacs mode.
Notebooks as Python scripts
When opening an existing notebook using the default settings, it is automatically converted to a Python script, with line comments of the form # %% defining cell boundaries. Additionally, the converted file begins with a YAML-like header block enclosed by two comment lines of the form # ---, which includes notebook metadata such as the kernel information as well as Jupytext format and version information. In order to execute cells, we can run M-x jupyter-repl-associate-buffer to associate the buffer to an existing emacs-jupyter REPL or create a new one choosing the appropriate Jupyter kernel. Cells with shell commands and IPython magics can be commented, which avoids syntax checking errors and allows the other IDE features to work properly14. An associated caveat is that such cells have to be uncommented before they can be evaluated by emacs-jupyter - otherwise we are just evaluating Python comments and thus seeing no effect whatsoever15. The following screencast illustrates how a notebook can be edited as Python script and its cells can be executed interactively using emacs-jupyter:
The fact that we are editing a Python file-like buffer has several key advantages when compared to the web-like interface of EIN, i.e., IDE features work fast and other basic features such as undo, auto saving and recovery work as in any regular Emacs file. However, in my view, notebooks are displayed more nicely in EIN: there is no need for a YAML-like header block16, cells feel more demarcated, markdown cells can be properly rendered instead of appearing as Python comments and most importantly, cell outputs (including plots) appear within the same notebook buffer. The latter brings me to the main shortcoming of this approach: as of May 2023, it is not possible to include the cell outputs in script representations of Jupyter notebooks17. As discussed in the introduction, this can be a crucial deficiency as it disables one of the major strengths of Jupyter notebooks, namely their storytelling ability. Without displaying cell outputs, notebooks are no longer such a suitable medium for tutorials, documentation or supporting materials of an academic article. Nonetheless, the combination of code-cells, Jupytext and emacs-jupyter can be very well suited to use cases where cell outputs can be ommited.
Finally, the configuration provided for this section of the post includes a couple of changes. First, I have found the default key bindings (all prefixed with C-c %) rather impractical, so the cell navigation, moving and execution key bindings are redefined to mimic EIN. Secondly, unlike EIN, code-cells and Jupytext do not provide any command to create a new notebook (with its metadata), so the configuration includes a function named my/new-notebook, which takes the new notebook file path and the desired Jupyter kernel as arguments.
Notebooks as org files
As its name suggests, org mode was initially created as an Emacs to organize notes and tasks. Nevertheless, what started lightweight markup language quickly evolved into a much more powerful system, notably thanks to Babel, a feature of org mode18 that allows users to include and execute source code blocks within org files. This is actually quite reminiscent of Jupyter notebook - in fact, one may argue that org and Babel provide a considerable superset of features, as they allow using multiple programming languages in the same file and offer richer and more flexible export capabilities.
It is actually very easy to configure code-cells to automatically convert notebooks to org files and edit them accordingly - provided that pandoc is installed, code-cells-convert-ipynb-style:
(setq code-cells-convert-ipynb-style '(
("pandoc" "--to" "ipynb" "--from" "org")
("pandoc" "--to" "org" "--from" "ipynb")
org-mode))
When opening the example notebook using the above configuration, the resulting org buffer converts Jupyter cells to org code blocks with the jupyter-python language identifier, with the markdown cells which are translated to org syntax (which start with a line of the form <<a0c84e00>> that corresponds to the Jupyter cell id). Like when editing notebooks as Python scripts, we need a running emacs-jupyter REPL to evaluate code blocks. However, to execute jupyter-python blocks in org mode, we also need a session, which is started by the Jupyter kernel and maintains the state, e.g., including any variables or objects that have been defined so that they can be shared between code blocks. Sessions can be defined using the session header argument, e.g.:
#+begin_src jupyter-python :session foo
import matplotlib.pyplot as plt
import rasterio as rio
from rasterio import plot
#+end_src
By default, emacs-jupyter uses the kernel associated with the currently active environment (see a dedicated section about this in the introduction above), yet this can be overridden by setting the :kernel header argument. Additionally, it is most certainly desirable to set the :async header argument to yes to allow asynchronous execution of code-blocks - otherwise the buffer will be completely stalled until the code execution is completed. Altogether, an example code block looks as follows:
#+begin_src jupyter-python :session foo :kernel jupyter-emacs :async yes
import matplotlib.pyplot as plt
import rasterio as rio
from rasterio import plot
#+end_src
While this highlights another advantage of org over Jupyter, namely the ability to use multiple languages and sessions in a single file, having to set these header arguments in each code block is quite verbose when only a single session and language is required - which surely corresponds to most use cases. Luckily, such a repetition can easily be avoided by setting the header-args property at the beginning of the file19:
#+PROPERTY: header-args:jupyter-python :session foo :kernel jupyter-emacs
#+PROPERTY: header-args:jupyter-python+ :async yes
so that all jupyter-python src blocks include the defined header arguments implicitly. Note that although it is beyond the scope of this post, it is further possible to set the session property to use remote kernels and much more20. In any case, once the session and the kernel have been properly configured, code blocks can be executed using the C-c C-c key bindings. The screencast below shows how the example notebook can be set up and executed as an org file buffer:
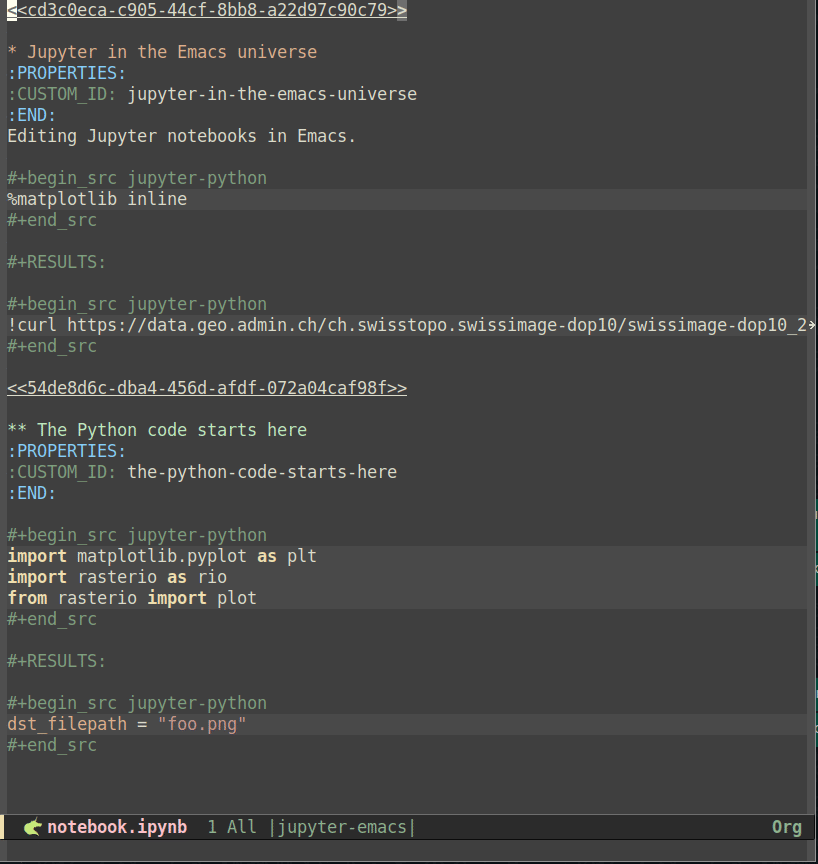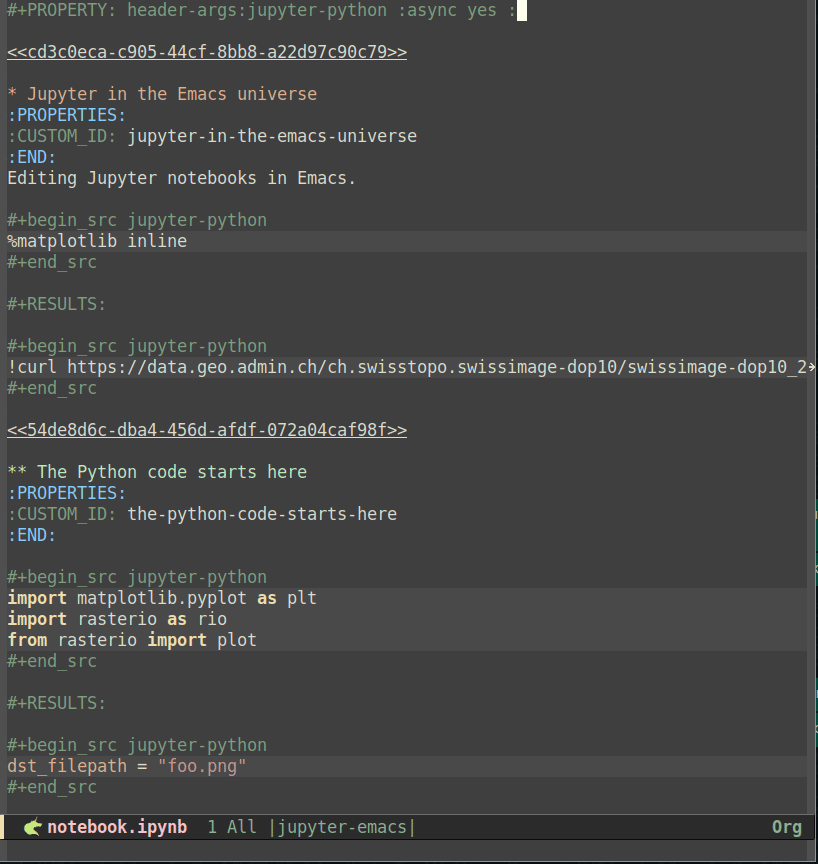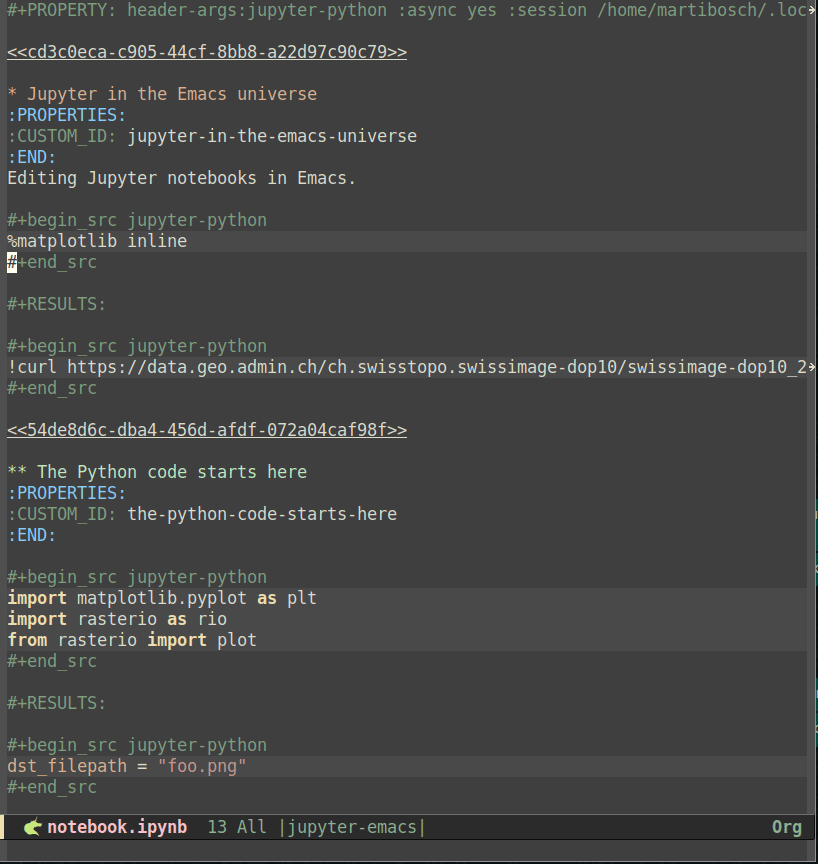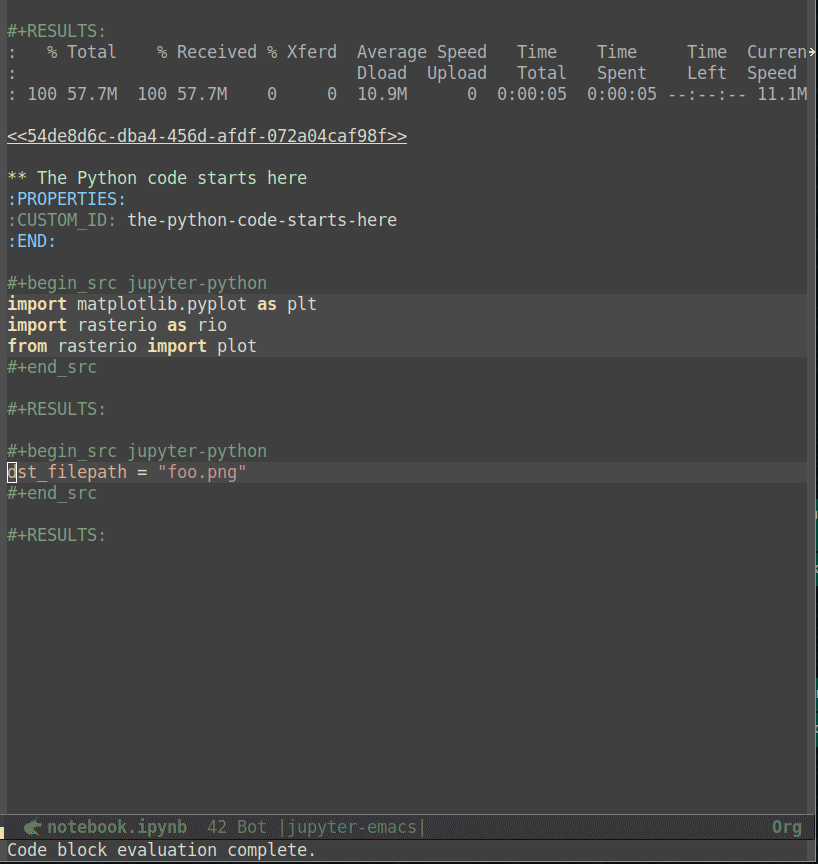
Likewise notebooks as Python scripts, org files are plain text, so the basic Emacs undo, auto saving and recovery work out of the box. Additionally, code blocks feel more demarcated than comment-delimited cells in Python scripts, and results can be displayed inline, including images - to that end, run org-toggle-inline-images (by default bound to C-c C-x C-v) after a code block producing an image has been executed21. Regarding key bindings, new code blocks (and other kinds of org mode blocks) can be inserted using the org-insert-structure-template command, by default bound to C-c C-,, and Babel provides a series of useful functions bound to the C-c C-v prefix that facilitate navigating to the next or previous code block (using C-c C-v n or C-c C-v p respectively) as well as to execute all code blocks in the buffer (bound to C-c C-v b). Even though I find these key bindings to be less handy than its counterparts in EIN and code-cells, this is understandable as org mode provides much more features and flexibility. Following the screencast above, the one below displays how code blocks can be edited and executed:
In my view, the main issue of using org to emulate Jupyter is related to how code blocks are edited. First, in order to get proper indentation and IDE features, code blocks must be edited in a separate buffer by using the org-edit-special command, by default bound to C-c '. But besides the inconvenience of having to use multiple buffers22 (especially in small screens), separate code editing buffers have a major drawback, i.e., they make getting proper IDE features excessively intricate (besides the default inspection and completion provided by emacs-jupyter). In fact, even after asking around in Reddit and GitHub issues I have not managed to achieve it23.
Finally, even though pandoc supports round-trip conversion from org to ipynb since version 2.19.2 (the org propertiy definitions at the start of the file are lost, but this is a minor detail), results from code blocks are ommited, hence the converted “.ipynb” files do not include cell outputs24.
Overall, org is nothing but adaptable so if you are comfortable tweaking your Emacs configuration, it is likely possible to suit org to your needs. In my experience, my inability to obtain proper notebook-wide IDE features when editing code is too much of a drawback.
A final note on polymode
Before wrapping up, it is worth noting that many of the problems in getting proper edition of Python code in org files is because the Python major mode is not active by default in org buffers, which makes sense because code blocks can be in many languages. Nonetheless, there exist libraries to support multiple major modes in a single buffer in Emacs - a notable example is polymode. It is possible to use code-cells with Jupytext or pandoc, transform notebooks to a plain-text representation such as markdown and then open them in a single polymode buffer so that fenced code blocks with a language identifier, e.g.:
```python
print("Here is some Python code")
```
can be edited in the language-specific major mode and sent to a running REPL. Nevertheless, doing so requires non-trival configuration based on a thorough understanding of how polymode works. Like with org mode, I have searched through blogs, GitHub and Reddit and I not been able to find any working configuration to make polymode work properly with emacs-jupyter or LSP25.
Conclusion
As one would expect, the choice of an appropriate setup largely depends on the use case. The key points to consider from the configurations reviewed in this post are summarized below.
TL;DR
Use EIN if you:
- want an interface as close as possible to Jupyter notebook and Jupyterlab, with a nice notebook display with code, rendered markdown and results
- want a simple Emacs setup that works out of the box but is inevitable less customizable
- do not mind slow IDE features (e.g., completion, documentation…)
- are going to be (very) careful not to mistakenly delete cells and save your notebooks constantly
Use notebooks as Python scripts (i.e., code-cells interacting with an emacs-jupyter kernel, with notebooks converted to Python scripts with Jupytext) if you:
- want all the advantages of working with plain text files (i.e., fast IDE features, undo, autosave…)
- do not mind a lightweight emulation of the Jupyter notebook experience, with outputs in separate buffers, lesser-demarcated cells and without rendered markdown
Use notebooks as org files (i.e., org-mode interacting with one or serveral emacs-jupyter kernels, with notebooks converted to org files with pandoc) if you:
- want a highly versatile and custumizable setup that largely supersedes the features offered by Juptyer (e.g., multiple kernels in the same file, checklists, agenda, calendar…), while keeping the advantages of working with plain text files
- are comfortable with hacky Emacs configurations and do not mind a steep learning curve (e.g., lots of metadata involved such as properties or header arguments, complex key bindings…)
Final (personal) thoughts
After much experimentation, my preference leans towards a setup where notebooks are by default edited as Python scripts (with code-cells, emacs-jupyter and jupytext as reviewed above), mainly because I find that the advantages of working with a plain-text file (i.e., fast IDE features, undo, autosave…) outweight the better notebook representation offered by EIN.
In my experience, the cell-centric feel provided by EIN and org-mode (as well as in Jupyter and Jupyterlab) ease the conceptual organization of code in a way that is not matched in code-cells, but this is merely a UI aspect that could be easily improved. On the other hand, I find the inability of the “notebooks as Python scripts” approach to include inline inputs17 to be a bit more troublesome, especially since it prevents including outputs in version control - again, hampering the storytelling ability of Jupyter notebooks. The latter is why my setup keeps the possibility of editing notebooks with EIN by running M-x ein:run or M-x ein:login and opening the notebooks from the EIN menu. I find this useful once the content of the notebook is solid and I just want to review it (e.g., follow its story, visualize outputs inline, proofread markdown…) before commiting it to version control. Actually, I often perform this stage in the browser so that I can see exactly what the notebook will look like in GitHub.
Finally, I have never needed to use more than one programming language or Jupyter kernel within the same file, therefore I have felt overwhelmed by org mode. But again, your needs may be different - in any case, I hope that this post provides a thorough and helpful overview of the strengths and weaknesses of each approach to write Jupyter notebooks within Emacs.
Notes
-
See Jake VanderPlas (2015). “The State of the Stack”, In 14th Python in Science Conference (SciPy). Slides. ↩
-
See Jeffrey M. Perkel (2018). “Why Jupyter is data scientists’ computational notebook of choice”, Nature. www.nature.com/articles/d41586-018-07196-1. ↩
-
See Jeffrey M. Perkel (2021). “Ten computer codes that transformed science”, Nature. www.nature.com/articles/d41586-021-00075-2. ↩
-
See Deutsch, L., E. Berkeley, and D. G. Bobrow (1964). “The LISP implementation for the PDP-1 computer.”. s3data.computerhistory.org/pdp-1/DEC.pdp_1.1964.102650371.pdf. ↩
-
See Rule, A., Birmingham, A., Zuniga, C., Altintas, I., Huang, S. C., Knight, R., … & Rose, P. W. (2019). Ten simple rules for writing and sharing computational analyses in Jupyter Notebooks. PLoS computational biology, 15(7), e1007007. doi.org/10.1371/journal.pcbi.1007007. ↩
-
I am thinking of the “I Don’t Like Notebooks” talk at JupyterCon 2018 by Joel Grus (link to the slides but there have been many others. Overall, I would use a quote from Reddit about the talk by Joel Grus to summarize the issue of notebooks, hidden state and reproducibility: “Notebooks are powerful, yes, but something can be said for mastering the screwdriver before diving into power tools.”. ↩
-
See “Python Programming in Emacs” at the EmacsWiki. ↩
-
The source code of my emacs setup is available at martibosch/snakemacs. ↩
-
To connect a given conda/mamba environment to Jupyter, you first need to activate the environment (i.e., run
conda activate <env-name>), install ipykernel as inconda install -c conda-forge ipykernel, and finally, the environment’s Jupyter kernel can be registered to the list of available kernels by runningpython -m ipykernel install --user --name=<env-name>. ↩ -
When it comes to managing virtual environments, I see two major advantages of conda/mamba, namely the ability of conda environments to manage the installation and update of Python itself (whereas virtualenv and venv rely on an existing and externally-managed Python executable - often the system Python itself) and manage non-Python packages, e.g., GDAL, CUDA dependencies and many more. See “Conda: Myths and Misconceptions” by Jake VanderPlas and “The definitive guide to Python virtual environments with conda” by WhiteBox. ↩
-
See “Create Virtual Environment using “virtualenv” and add it to Jupyter Notebook” for an example alternative (to conda) setup for Python virtual environments and Jupyter using virtualenv. It should be possible to further enable automatic environment activation in Emacs using auto-virtualenv or poetry.el. ↩
-
A caveat of the proposed conda and emacs setup is that pyright needs to be installed within each conda environment - otherwise the static type checking will not work when editing an Emacs buffer with an active conda environment. The same holds for formatting and linting tools such as black or ruff. To ensure that the required packages are installed in all conda environments by default, you may use the
create_default_packagesoption in your.condarcconfiguration file. See the “Conda environments and IDE features for Python buffers” section of the snakemacsREADME.mdfor a more detailed overview of the caveat. ↩ -
See github.com/millejoh/emacs-ipython-notebook/issues/841. ↩
-
The completion at point mechanisms from emacs-jupyter actually override those from LSP. Ideally, it should be possible to disable the emacs-jupyter completion in Python buffers so that all the IDE features are provided by LSP. See github.com/nnicandro/emacs-jupyter/issues/344. ↩
-
In fact, as of May 2023 there is another related bug, which is that evaluating cells with cell-level magics using the
code-cells-evalcommand raises a JupyterUsageError. Instead, cell-level magics work properly when selecting the cell region manually and usingjupyter-eval-region. See github.com/astoff/code-cells.el/issues/18. ↩ -
It should be noted that the header block can be reduced to only include the kernel information by setting the Jupytext metadata in a configuration file. See the “Jupytext configuration file” of the Jupytext documentation. ↩
-
There is actually a proposal by Marc Wouts (the author of jupytext) to support including outputs in the percent format of Jupyter notebooks. ↩ ↩2
-
Babel was initially an extension to org, but it is integrated into the org core since its version 7.0. See the org Manual. ↩
-
In the provided snippet, the property has been defined in two lines using the
+sign to avoid exceeding certain line length, but it could have been equivalently defined in a single line. ↩ -
See the excellent post “Replacing Jupyter Notebook with Org Mode”, by Pavel Korytov. ↩
-
It is a bit hacky, but it is possible to configure org to display inline images automatically after execution of a respective code block by adding the following hook:
(add-hook 'org-babel-after-execute-hook 'org-display-inline-images 'append). See the “Tips and tricks” section of the README file ofob-ipython. ↩ -
Furthermore, the tab and return keys are mapped to
org-cycleandorg-returnrespectively, which have quite a complex behaviour that is very tricky to configure. This can be disabled by settingorg-src-tab-acts-nativelytonil, but then the code is not automatically indented. There is quite a vast collection of threads in the Emacs sub-reddit and questions in the Emacs Stack Exchange about this issue, which in my experience all discourage inline editing in org mode. ↩ -
While I have managed to obtain buffer-wide IDE features with
org-edit-special, such a buffer corresponds to a single code block (equivalent to a single notebook cell). Therefore, the challenge is to obtain proper IDE features based on the whole org file (equivalent to the whole notebook), e.g., completion considering variables defined in other code blocks. In org mode, this would require tangling code blocks to a file and activating an LSP client. Nevertheless, this is not yet officially documented in lsp-mode, and the dedicated lsp-org mode is still in an experimental/alpha phase. Similarly, this remains an open issue in eglot. Additionally, tangling should be automated and synchronized (e.g., with tools such as org-tanglesync or org-auto-tangle) so that IDE features are updated as source blocks are edited. Finally, I have seen suggested the noweb syntax may be used so that source code blocks can include references to other code blocks. However, as mentioned in the post above, I have not been able to find any working configuration that puts it all together to obtain org file-wide/notebook-wide IDE features. ↩ -
The ox-ipynb module allows exporting org files to ipynb while preserving cell outputs. The tool does not support round trip conversion, but it should be possible to use ox-ipynb to export to ipynb, and then pandoc to convert them back to org if the file needs to be edited again. However such a is not straightforward to configure using code-cells, as
code-cells-convert-ipynb-styleexpects strings representing shell commands and ox-ipynb does not provide any - it provides Emacs commands only. ↩ -
I have not managed to find any configuration that properly links emacs-jupyter with polymode. While it is possible to use emacs-jupyter commands such as
jupyter-eval-line-or-regionin Python code blocks of polymode buffers, by default these commands do not recognize the code block boundaries, and thus the code has to be evaluated line by line. This can possibly be achieved but doing so requires a comprehensive understanding of how polymode works, which at first glance I have found to be too demanding. On the other hand, regarding IDE features, I managed to make LSP work following an approach proposed in a GitHub issue, but it was quite impractical in terms of performance as LSP was activated at each code block, therefore, to get environment-aware IDE features, this also requires activating the corresponding environment. Similarly to org mode, this allows you to use multiple languages in the same notebook-like file, yet I rarely - or practically never - need that. ↩
